Ontogenesis and molecular interaction
Context
Proteins are the universal elements of life. They form tightly interconnected networks called “interactomes” to ensure their diverse and specific functions in the cell. Deciphering interactomes is a key issue in biology for understanding fundamental aspects of life, but also for cracking the molecular codes deregulated in human diseases.
Our team is tackling the issue of interactomes established by evolutionary conserved transcription factors, such as the Hox proteins. Hox proteins are major regulators of embryonic development in animals, specifying cell fate in all tissues from early to late developmental stages. Hox proteins are also required in the adult, in particular for regulating stem cell populations in different tissues. In contrast to the diverse and specific roles of Hox proteins, their interaction partners remain poorly characterized. More generally, the capture of interactomes remains a challenging issue in living cell contexts.
Our team has established and is developing tools to visualize and capture protein-protein interactions in living Drosophila and human cells at various resolution scales. Our approaches cover genetics, microscopy, proteomics and modelling to unravel new paradigms underlying the formation and activity of interactomes.
Key words
Protein-protein interaction, gene regulation, Drosophila, human cells, complementation, Hox
METHODS
Research projects are based on Bimolecular Fluorescence Complementation (BiFC), which has been further implemented for performing large-scale interaction screens in Drosophila (Bischof et al., eLife 2018) and human cells (Jia et al., Cells 2024). We also established Photo-Activated BiFC for visualizing protein-protein interactions at the single molecule resolution level (Vanderperre et al., Springer Protocols book, in press). Finally, we recently developed new tools for testing the impact of drugs on specific interactomes (Kundlacz et al., in preparation), and established a novel methodology for capturing endogenous interactomes of specific dimeric protein complexes in human cells (Hajj Sleiman et al., BioRrxiv 2024).
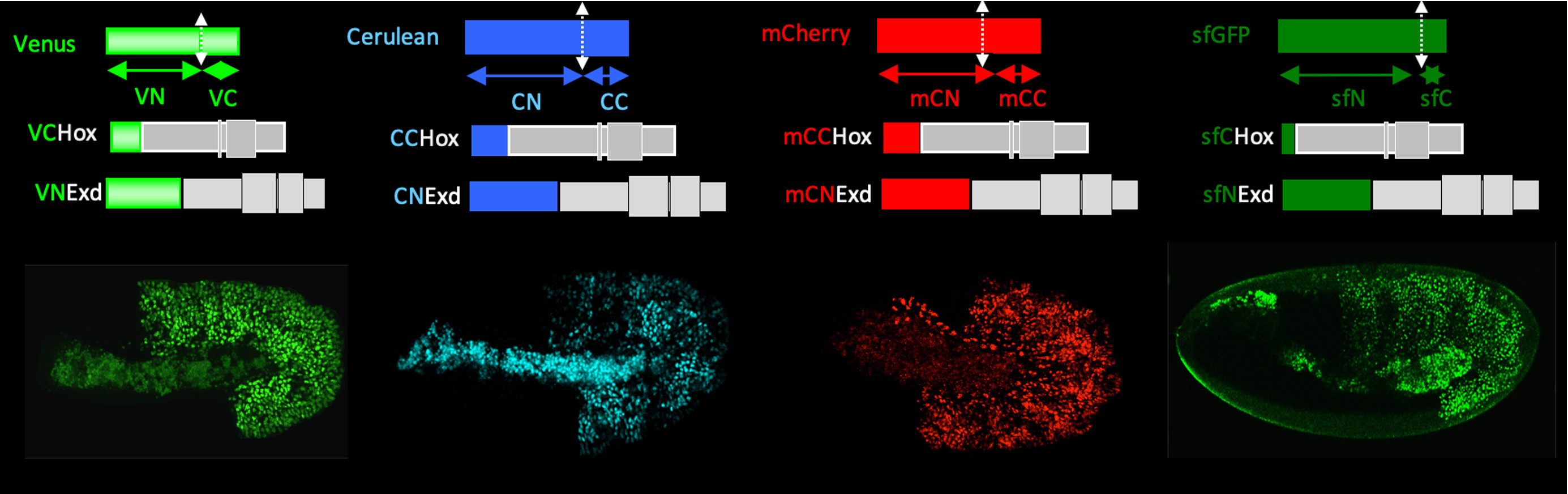
BiFC with different fluorescent proteins in living drosophila embryos
research projects
Our research projects use the Hox proteins in different biological contexts as model systems to tackle the issue of interactome specificity.
1. Role of LaminC and Hox proteins in autophagy repression during Drosophila development
This project relates to a novel partnership between Hox proteins and the LaminC nuclear component for repressing autophagy in the Drosophila larval fat body. This project aims at (i) characterizing the target genes and the interactomes of repressive Hox/LamC complexes and (ii) deciphering protein interaction dynamics at the super resolution scale in fat body nuclei.
2. Role of LaminC and Hox proteins in autophagy repression for maintaining homeostasis in the Drosophila adult fly
This project aims at understanding whether the molecular cues underlying Hox/LamC repressive activity on autophagy are identical or not between the larval and adult stages. Since autophagy activity declines with age, this project also aims at characterizing the role of Hox and Lamin C proteins in regulating ageing and age-associated diseases during the adult life.
3. Hox and LaminC as a novel partnership for preventing adipogenesis and promoting osteogenesis
Hox proteins repress adipogenesis, and promote osteogenesis with the help of the RUNX2 transcription factor. A-type Lamins are also known to repress adipogenesis and we postulate that this repressive activity could rely on a partnership with Hox proteins, as observed in Drosophila. This project aims at validating this hypothesis, and more globally at identifying Hox interactomes underlying their opposite functions for adipogenesis and osteogenesis.
4. Technological development
Our projects rely on the development of advanced tools for getting access to functionally relevant interactomes and their molecular dynamics in live conditions.
4.1. Improving tools for capturing interactomes of binary protein complexes in living cells
Our recent work established Bi-nano-ID as a novel methodology for capturing endogenous interactomes of dimeric protein complexes (Hajj Sleiman et al., BioXriv 2024). Our approach is based on a nanobody recognizing BiFC complexes and we aim at further improving the specificity of this tool by testing alternative nanobodies.
4.2. Establishing splitFAST in Drosophila to visualize protein interaction dynamics in living embryos
This project aims at implementing splitFAST in the Drosophila embryo. This cutting-edge methodology allows visualizing protein-protein interaction dynamics and is based on the property of the FAST enzyme to be reassembled upon spatial proximity (Rakotoarison et al., ACS Chem Biol 2024). This project is performed in collaboration with the group of A. Gautier (Sorbonne University, Paris, France).
HOW TO CONTACT US
We welcome students for internships, don’t hesitate to contact Samir Merabet (samir.merabet@ens-lyon.fr) if you are interested!


