Thesis defense: Ziyan Nie
| When |
Nov 13, 2025
from 02:00 to 05:00 |
|---|---|
| Where | Salle des thèses |
| Contact Name | Ziyan Nie |
| Attendees |
Florence BESSE, Université Côte d'Azur / CNRS Rapporteure; Florian MULLER, Institut Pasteur Rapporteur; Zayna CHAKER, IGFL; Davide NORMANNO, Université de Montpellier; Jean LIVET, Sorbonne Université / INSERM; Jonathan ENRIQUEZ, IGFL, Directeur de thèse. |
| Add event to calendar |
|
On November 13th, Ziyan Nie of the team of Jonathan Enriquez will support his thesis entitled:
"The role of Imp and Syp RNA-binding proteins in the precise control of motor neuron number in a single neural stem cell lineage, and development of a MERFISH platform for Drosophila spatial transcriptomics"
Abstract:
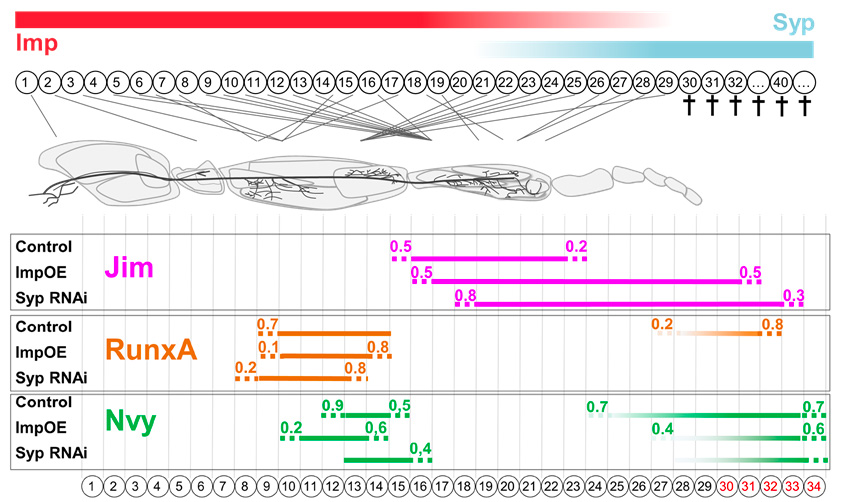
This thesis project utilizes Drosophila melanogaster to dissect the mechanisms governing precise and consistent neuron number. I investigated how neural stem cells in the ventral nerve cord, known as neuroblasts (NBs), control progeny number and, beyond NB activity, how the fate of immature motor neurons (iMNs) plays a crucial role. Specifically, within the Drosophila Lin A/15 neuroblast lineage, which produces motor neurons and glia, approximately 40% of iMNs undergo apoptosis. With colleagues we identified two RNA-binding proteins, Imp and Syp, as key regulators: Imp expression correlates with iMN survival, while Imp-negative, Syp-positive iMNs undergo apoptosis. My findings revealed that normally eliminated late-born iMNs fail to express the correct combinatorial code of transcription factors (mTFs) specifying early-born MN identity. Crucially, manipulating Imp and Syp in the Lin A/15 lineage altered the mTF profile of last-born iMNs to resemble earlier-born MNs, promoting their survival. Furthermore, genetically imposing the earlier-born MNs' mTF code was sufficient to promote iMN survival. These results demonstrate a molecular mechanism where the temporally differential expression of Imp and Syp in iMNs couples precise neuronal generation with identity acquisition by regulating fate-determining mTFs, suggesting a fundamental, conserved mechanism in neurogenesis.
Building on some of the methods applied in the first part, my project subsequently transitioned to implementing imaging-based spatial transcriptomics, specifically Multiplexed Error-Robust Fluorescence in situ Hybridization (MERFISH), with the aim of spatially and quantitatively profiling transcription factors and other signature genes in Drosophila leg imaginal discs at single-cell resolution. In addition, these efforts also contributed to the implementation of a versatile and species-agnostic spatial omics platform.