Thèse: Domitille CHALOPIN
| Quand ? |
Le 23/05/2014, de 09:00 à 12:30 |
|---|---|
| S'adresser à | Domitille Chalopin |
| Ajouter un événement au calendrier |
|
Domitille CHALOPIN de l'équipe de J.-N. Volffsoutiendra sa thèse (en anglais) le vendredi 23 mai à 09h00 dans l'amphithéâtre Schröedinger (ENS, site Monod).
Sa thèse s'intitule:
"Comparative genomics of transposable element evolution and their evolutionary impacts in fish and other vertebrate genomes"
Résumé:
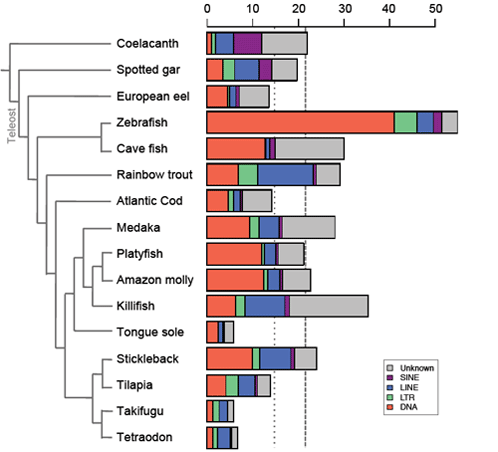
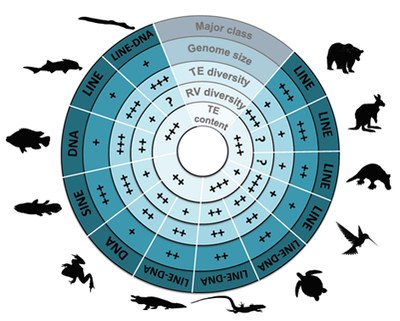
| Transposable elements (TEs) are mobile genetic elements - able to move and to multiply within genomes - identified in almost all living organisms including bacteria. Considered as junk DNA for long, nowadays they are undeniably major players of gene, genome and host evolution. TEs can be deleterious causing diseases at the individual scale, but they are strong evolutionary agents involved in genome plasticity at a larger scale. Furthermore, these “parasites” can also be source of new genetic materials as promoters or even new genes bringing new functions for hosts. The main objectives of my thesis was to determine the presence or not of the different TE families in fish genomes, as well as their respective content to understand the evolutionary history of these families in fish genomes compared to other vertebrate genomes. Being involved in various genome sequencing consortium projects (including the coelacanth one), I performed a large-scale comparative analysis to highlight the various evolutionary strategies of TEs. I showed that TE content is highly variable in vertebrate genomes, the smallest and the largest being found in fish (6% in tetraodon and 55% in zebrafish), and may contribute to their genome sizes especially in fish that present more TEs than compared sarcopterygian genomes of same sizes. Analyses of TE superfamilies also present a large variation between species. While some superfamilies are widespread (ERV, Penelope, L1, CR1-like retrotransposons, Tc-Mariner, hAT transposons), some have been completely lost in particular lineages, such as CR1 in teleost fish, and others present very patchy distribution, as Merlin, also suggesting that several events of horizontal transfer occurred. These superfamilies underwent differential waves of activity in vertebrate species highlighting TE dynamics and predicting the potential current activity in each genome. On another hand, I particularly focused on the study of a vertebrate-specific TE-derived gene, named Gin-2, to understand its origin, evolution, and its potential function in vertebrates that is completely unknown. In silico analyses showed that Gin-2 is a very ancient gene (around 450 My) deriving from a GIN transposon and is absent from placental mammals. Further analyses present a particular expression in brain and gonads during adulthood, while a strong expression during gastrulation suggests a potential role of Gin-2 in zebrafish development. All together, the different analyses performed during my thesis contribute to a better view of TE evolution and their evolutionary impacts in vertebrate genomes (content, diversity, dynamics, activity, exaptation, functions…). | |||
 |
 |
||


