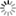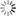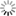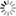mothur fig4
#! /Users/mtefit/Bioinfo/mothur/mothur
system(cd /Users/directoryname)
#create master fasta file + corresponding group file
make.group(fasta=/Users/directoryname/filename_AB1.fasta-/Users/directoryname/filename_A1.fasta-/Users/directoryname/filename_A2.fasta-/Users/directoryname/filename_A3.fasta-/Users/directoryname/filename_A10.fasta-/Users/directoryname/filename_A11.fasta-/Users/directoryname/filename_A12.fasta-/Users/directoryname/filename_A8.fasta-/Users/directoryname/filename_AB2.fasta-/Users/directoryname/filename_TPCR.fasta-/Users/directoryname/filename_TEXT.fasta-/Users/directoryname/filename_TLAB.fasta-/Users/directoryname/filename_A9.fasta-/Users/directoryname/filename_AB3.fasta-/Users/directoryname/filename_B4.fasta-/Users/directoryname/filename_AB4.fasta-/Users/directoryname/filename_B5.fasta-/Users/directoryname/filename_B6.fasta-/Users/directoryname/filename_A4.fasta-/Users/directoryname/filename_B7.fasta-/Users/directoryname/filename_A5.fasta-/Users/directoryname/filename_B8.fasta-/Users/directoryname/filename_A6.fasta-/Users/directoryname/filename_B9.fasta-/Users/directoryname/filename_B1.fasta-/Users/directoryname/filename_B2.fasta-/Users/directoryname/filename_B3.fasta-/Users/directoryname/filename_B10.fasta-/Users/directoryname/filename_B11.fasta-/Users/directoryname/filename_B12.fasta-/Users/directoryname/filename_A7.fasta, groups=AB1-A1-A2-A3-A10-A11-A12-A8-AB2-TPCR-TEXT-TLAB-A9-AB3-B4-AB4-B5-B6-A4-B7-A5-B8-A6-B9-B1-B2-B3-B10-B11-B12-A7)
system(mv /Users/directoryname/filename_mergegroups /Users/directoryname/filename_mergedsamples.group)
merge.files(input=/Users/directoryname/filename_AB1.fasta-/Users/directoryname/filename_A1.fasta-/Users/directoryname/filename_A2.fasta-/Users/directoryname/filename_A3.fasta-/Users/directoryname/filename_A10.fasta-/Users/directoryname/filename_A11.fasta-/Users/directoryname/filename_A12.fasta-/Users/directoryname/filename_A8.fasta-/Users/directoryname/filename_AB2.fasta-/Users/directoryname/filename_TPCR.fasta-/Users/directoryname/filename_TEXT.fasta-/Users/directoryname/filename_TLAB.fasta-/Users/directoryname/filename_A9.fasta-/Users/directoryname/filename_AB3.fasta-/Users/directoryname/filename_B4.fasta-/Users/directoryname/filename_AB4.fasta-/Users/directoryname/filename_B5.fasta-/Users/directoryname/filename_B6.fasta-/Users/directoryname/filename_A4.fasta-/Users/directoryname/filename_B7.fasta-/Users/directoryname/filename_A5.fasta-/Users/directoryname/filename_B8.fasta-/Users/directoryname/filename_A6.fasta-/Users/directoryname/filename_B9.fasta-/Users/directoryname/filename_B1.fasta-/Users/directoryname/filename_B2.fasta-/Users/directoryname/filename_B3.fasta-/Users/directoryname/filename_B10.fasta-/Users/directoryname/filename_B11.fasta-/Users/directoryname/filename_B12.fasta-/Users/directoryname/filename_A7.fasta, output=/Users/directoryname/filename_mergedsamples.fasta)
#sequence processing
summary.seqs(fasta=/Users/directoryname/filename_mergedsamples.fasta)
align.seqs(fasta=current, template=/Users/directoryname/Silva.nr_v123/silva.nr_v123.align, flip=T)
screen.seqs(fasta=/Users/directoryname/filename_mergedsamples.align, group=/Users/directoryname/filename_mergedsamples.group, optimize=start-end, criteria=90)
filter.seqs(fasta=current, vertical=T, trump=.)
unique.seqs()
pre.cluster()
chimera.uchime(fasta=current, name=current)
remove.seqs(fasta=current, name=current, group=/Users/directoryname/filename_mergedsamples.group)
classify.seqs(fasta=current, name=current, group=current, template=/Users/directoryname/Silva.nr_v123/silva.nr_v123.align, taxonomy=/Users/directoryname/Silva.nr_v123/silva.nr_v123.tax, cutoff=80)